Recent projects
Respiratory Syncytial Virus Genomics
Screen clinically relevant genomic variations of RSV from NGS data
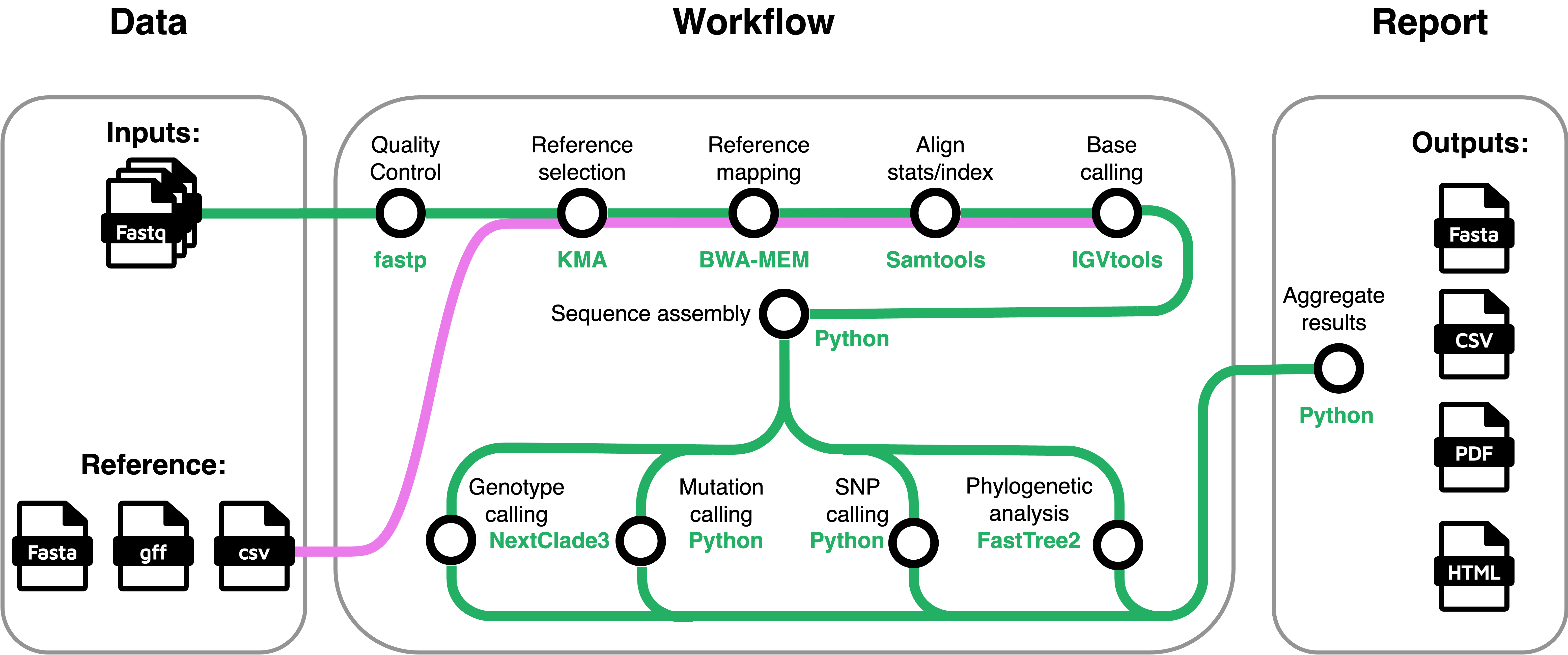
Github page: RSVreconPy and RSVrecon Paper: Wiley
Spatial Omics
Improving Data Interpretation for High Resolution Spatial Omics using computational approaches
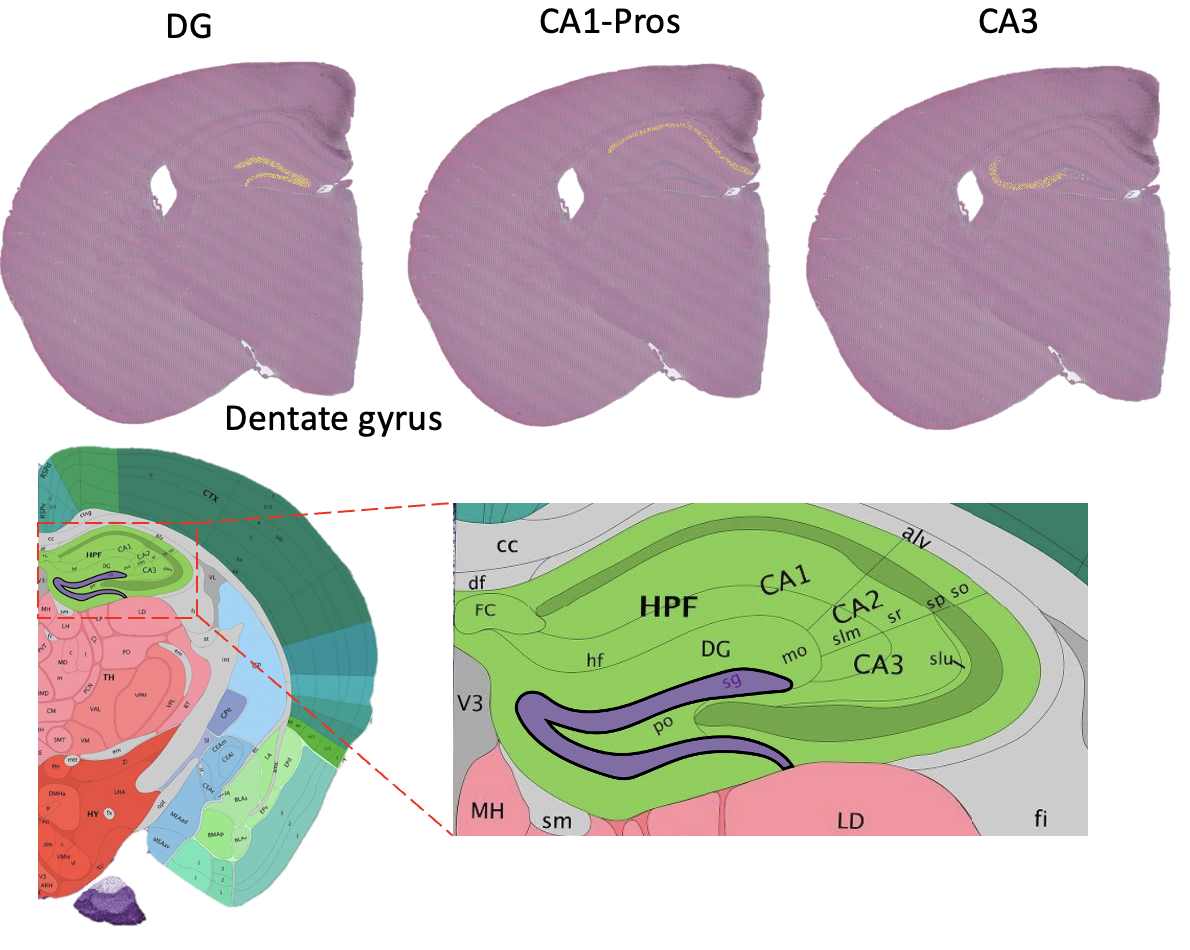
Github page: In preparation Paper: In preparation
Hydrid demultiplexing
A hybrid single cell demultiplexing strategy that increases both cell recovery rate and calling accuracy
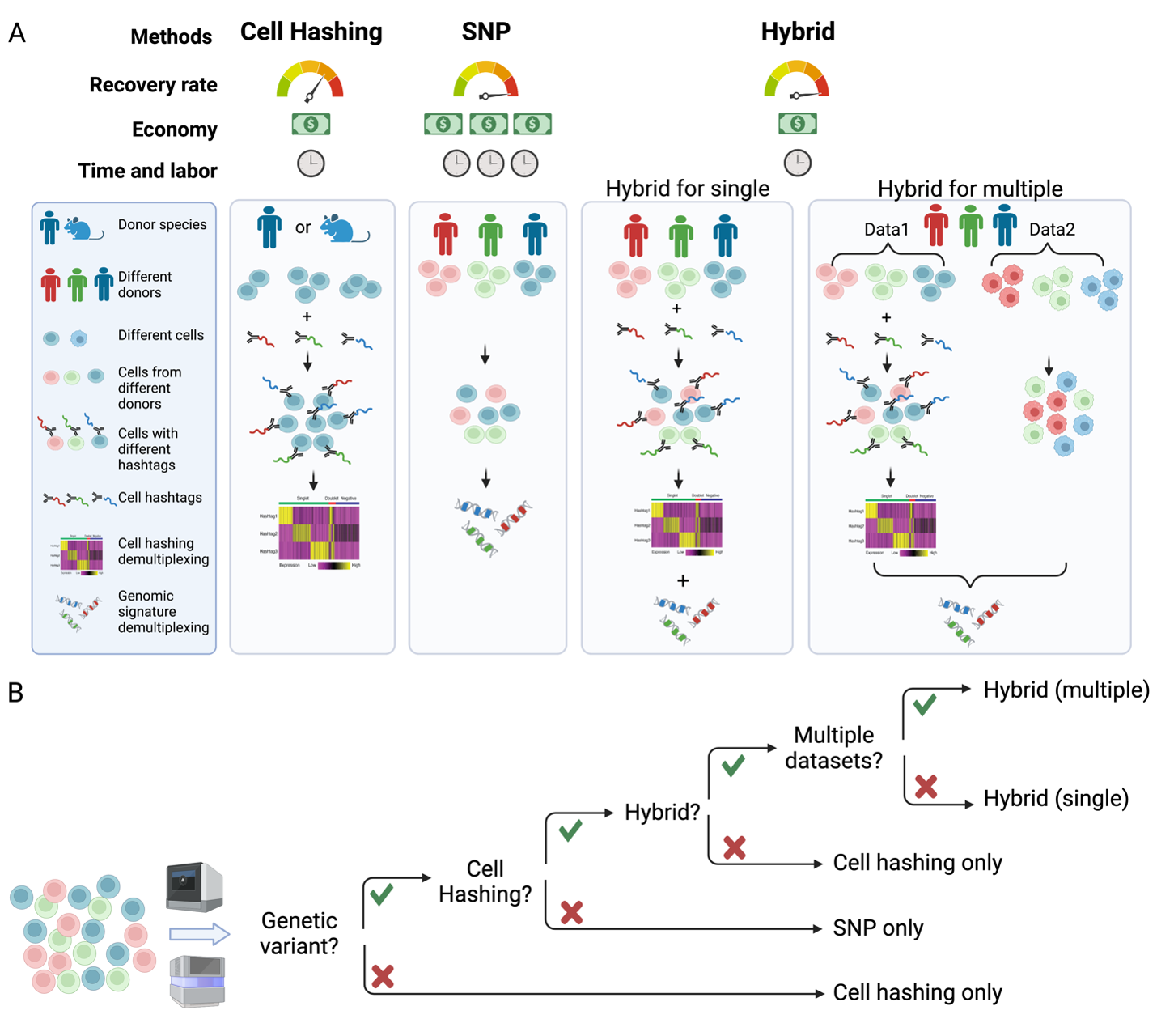
Github page: HTOreader Paper: Oxford Press
VGenes
VGenes is an integrated graphical tool for efficient, comprehensive and multimodal analyses of massive B-cell repertoire sequences
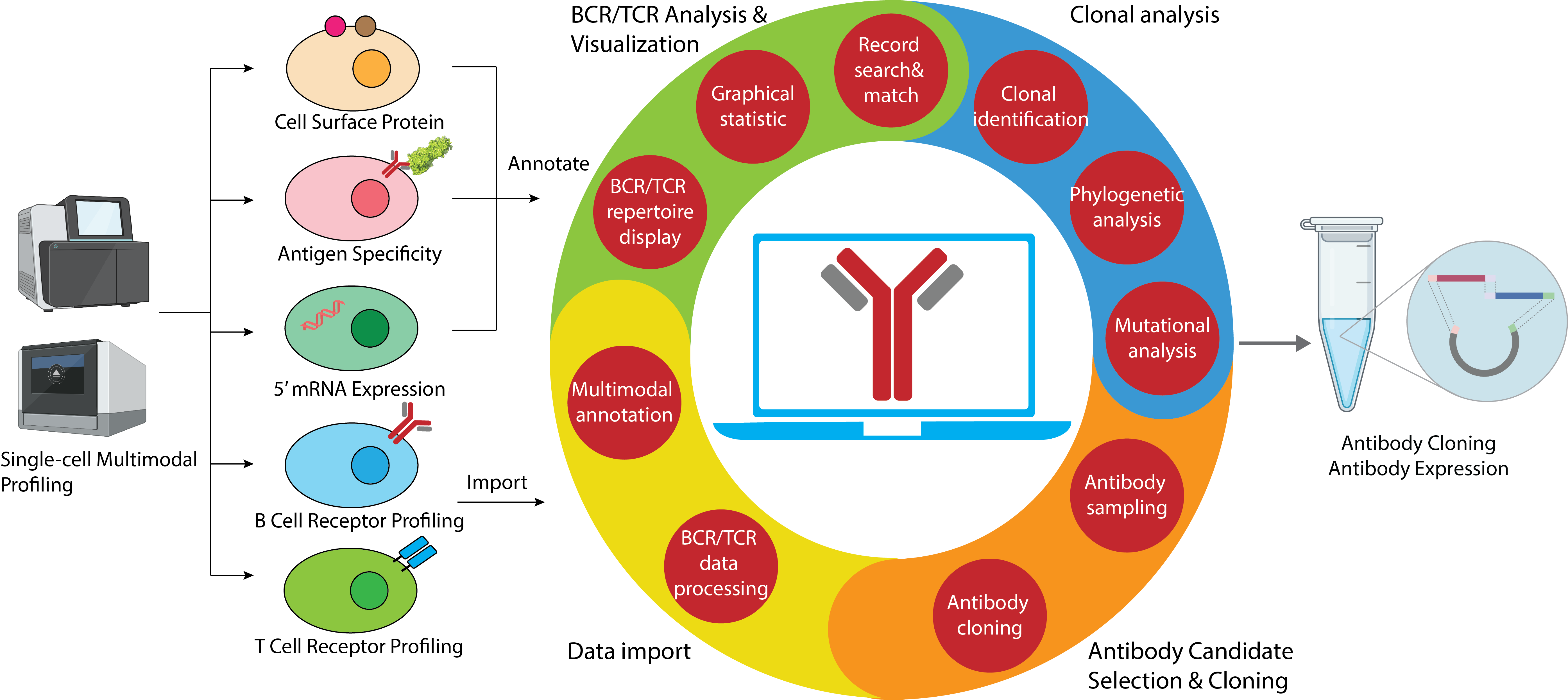
Github page: VGenes Paper: In preparation
Librator
Librator is a platform for optimized sequence editing, design, and expression of influenza virus proteins.

Github page: Librator Paper: Oxford Press
LinQ-View
LinQ-View is a single cell analysis strategy that could integrate RNA and ADT profiles for cell heterogeneity identification.
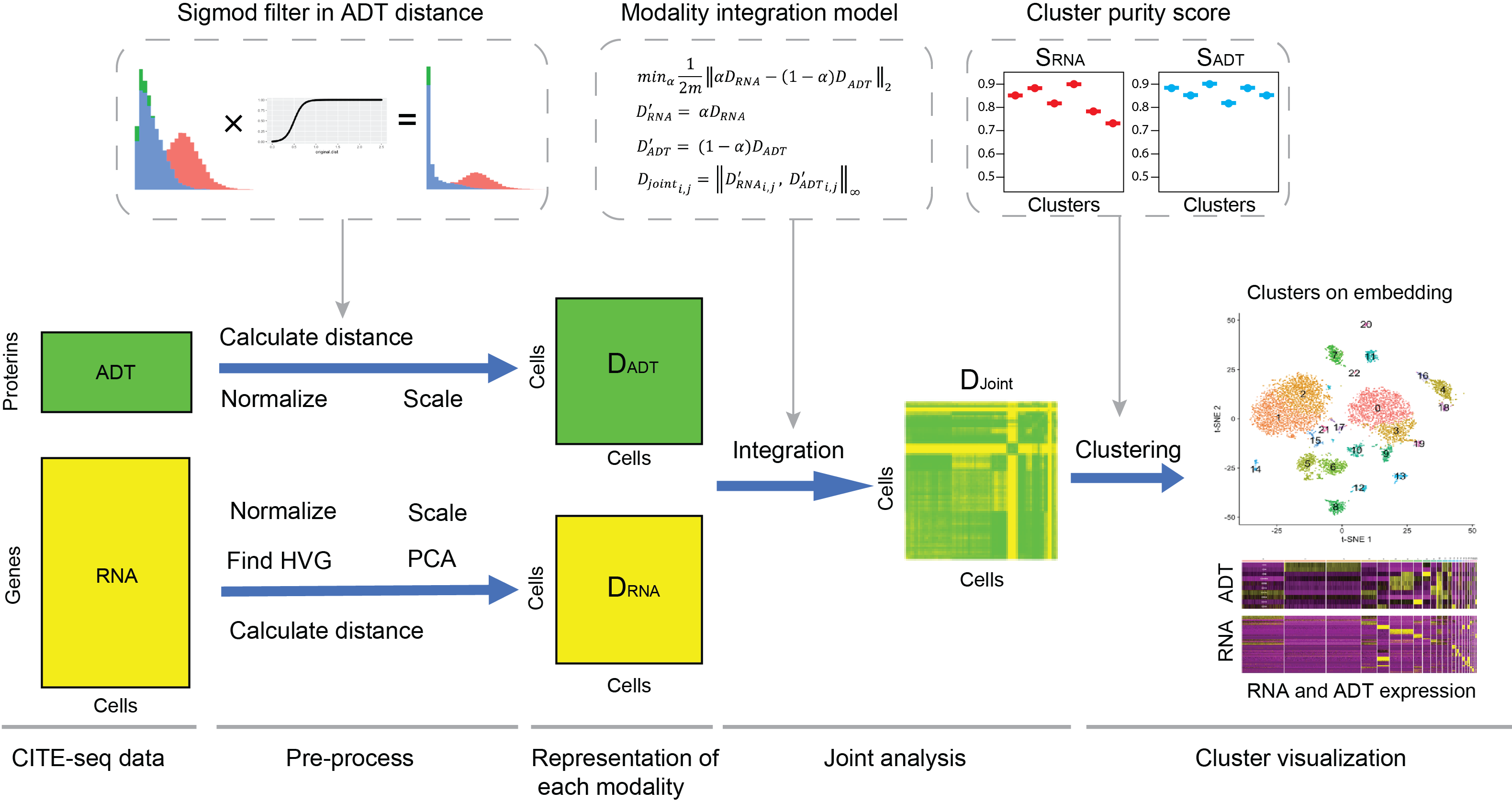
Github page: LinQ-View Paper: Cell Press
Cookie
Cookie: Selecting representative samples from single cell atlas using k-medoids clustering.

Github page: Cookie Paper: Frontiers in Genetics
Past projects
Dissecting the evolution of B cells against SARS-CoV-2 and Influenza viruses
- #Dugan, Haley L; #Stamper, Christopher; #Li, Lei; Changrob, Siriruk; Asby, Nicholas; Halfmann, Peter; Zheng, Nai-Ying; Huang, Min; Shaw, Dustin G; Cobb, Mari; Profiling B cell immunodominance after SARS-CoV-2 infection reveals antibody evolution to non-neutralizing viral targets Immunity 54, no. 6 (2021): 1290-1303. ScienceDirect
- #Li, Lei; #Dugan, Haley L; #Stamper, Christopher; Lan, Linda Yu-Ling; Asby, Nicholas; Knight, Matthew; Stovicek, Olivia; Zheng, Nai-Ying; Madariaga, Maria Lucia; Shanmugarajah, Kumaran; Improved integration of single-cell transcriptome and surface protein expression by LinQ-View Cell Reports Methods 1 (4), 100056 (2021) Cell Press
- Guthmiller, Jenna J; Utset, Henry A; Henry, Carole; Li, Lei; Zheng, Nai-Ying; Sun, Weina; Vieira, Marcos C; Zost, Seth; Huang, Min; Hensley, Scott E; An egg-derived sulfated N-Acetyllactosamine glycan is an antigenic decoy of influenza virus vaccines Mbio 12, no. 3 (2021): e00838-21. ASM Journals
- Siriruk Changrob, Yanbin Fu, Jenna Guthmiller, Peter Halfmann, Lei Li, Christopher Stamper, Haley Dugan, Molly Accola, William Rehrauer, Nai-Ying Zheng, Min Huang, Jiaolong Wang, Steven Erickson, Henry Utset, Hortencia Graves, Fatima Amanat, D Noah Sather, Florian Krammer, Yoshihiro Kawaoka, Patrick Wilson; Cross neutralization of emerging SARS-CoV-2 variants of concern by antibodies targeting distinct epitopes on spike ResearchSquare 2021 Preprint
- Guthmiller, Jenna J; Stovicek, Olivia; Wang, Jiaolong; Changrob, Siriruk; Li, Lei; Halfmann, Peter; Zheng, Nai-Ying; Utset, Henry; Stamper, Christopher T; Dugan, Haley L; SARS-CoV-2 infection severity is linked to superior humoral immunity against the spike MBio 12, no. 1 (2021): e02940-20 JSM
- Guthmiller, Jenna J; Han, Julianna; Li, Lei; Freyn, Alec W; Liu, Sean TH; Stovicek, Olivia; Stamper, Christopher; Dugan, Haley L; Tepora, Micah E; Bitar, Dalia J; First exposure to the pandemic H1N1 virus induced broadly neutralizing antibodies targeting hemagglutinin head epitopes Science Translational Medicine 2021 Vol. 13, Issue 596, eabg4535. DOI: 10.1126/scitranslmed.abg4535 Science Translational Medicine
- Knight, Matthew; Changrob, Siriruk; Li, Lei; Wilson, Patrick C; Imprinting, immunodominance, and other impediments to generating broad influenza immunity Immunological Reviews 2020 Wiley
- Guthmiller, Jenna J; Lan, Linda Yu-Ling; Fernández-Quintero, Monica L; Han, Julianna; Utset, Henry A; Bitar, Dalia J; Hamel, Natalie J; Stovicek, Olivia; Li, Lei; Tepora, Micah; Polyreactive Broadly Neutralizing B cells Are Selected to Provide Defense against Pandemic Threat Influenza Viruses Immunity 2020 Pubmed
- Krammer, Florian; Li, Lei; Wilson, Patrick C; Emerging from the shadow of hemagglutinin: neuraminidase is an important target for influenza vaccination Cell host & microbe 2019 Pubmed
Performing computational biology research to understand influenza genotype-phenotype corrolations
- Li, Lei; Chang, Deborah; Han, Lei; Zhang, Xiaojian; Zaia, Joseph; Wan, Xiu-Feng; Multi-task learning sparse group lasso: a method for quantifying antigenicity of influenza A (H1N1) virus using mutations and variations in glycosylation of Hemagglutinin BMC bioinformatics 2020 BMC
- Han, Lei; Li, Lei; Wen, Feng; Zhong, Lei; Zhang, Tong; Wan, Xiu-Feng; Graph-guided multi-task sparse learning model: a method for identifying antigenic variants of influenza A (H3N2) virus Bioinformatics 2019 PMC
- Martin, Brigitte E; Bowman, Andrew S; Li, Lei; Nolting, Jacqueline M; Smith, David R; Hanson, Larry A; Wan, Xiu-Feng; Detection of antigenic variants of subtype H3 swine influenza A viruses from clinical samples Journal of clinical microbiology 2017 Pubmed
Investigating origins and evolutionary pathways for avian influenza A (H5 and H7) viruses among domestic poultry
- Li, Lei; Bowman, Andrew S; DeLiberto, Thomas J; Killian, Mary L; Krauss, Scott; Nolting, Jacqueline M; Torchetti, Mia Kim; Ramey, Andrew M; Reeves, Andrew B; Stallknecht, David E; Genetic evidence supports sporadic and independent introductions of subtype H5 low-pathogenic avian influenza A viruses from wild birds to domestic poultry in North America Journal of virology 2018 JVI
- Li, Lei; DeLiberto, Thomas J; Killian, Mary L; Torchetti, Mia K; Wan, Xiu-Feng; Evolutionary pathway for the 2017 emergence of a novel highly pathogenic avian influenza A (H7N9) virus among domestic poultry in Tennessee, United States Virology 2018 Pubmed
- #Zhang, Xiaojian; #Sun, Hailiang; #Cunningham, Fred L; #Li, Lei; Hanson-Dorr, Katie; Hopken, Matthew W; Cooley, Jim; Long, Li-Ping; Baroch, John A; Li, Tao; Tissue tropisms opt for transmissible reassortants during avian and swine influenza A virus co-infection in swine PLoS Pathogens 2018 PLoS Pathogen
- Lee, Dong-Hun, Mary Lea Killian, Thomas J. Deliberto, Xiu-Feng Wan, Li Lei, David E. Swayne, and Mia Kim Torchetti. H7N1 Low Pathogenicity Avian Influenza Viruses in Poultry in the United States During 2018. Avian Diseases 65, no. 1 (2021): 59-62. Avian Diseases
Imprinting of repeated influenza A/H3 exposures on antibody quantity and antibody quality
- #Kosikova, Martina; #Li, Lei; Radvak, Peter; Ye, Zhiping; Wan, Xiu-Feng; Xie, Hang; Imprinting of repeated influenza A/H3 exposures on antibody quantity and antibody quality: implications for seasonal vaccine strain selection and vaccine performance Clinical infectious diseases 2018 Pubmed
- Xie, Hang; Li, Lei; Ye, Zhiping; Li, Xing; Plant, Ewan P; Zoueva, Olga; Zhao, Yangqing; Jing, Xianghong; Lin, Zhengshi; Kawano, Toshiaki; Differential effects of prior influenza exposures on H3N2 cross-reactivity of human postvaccination sera Clinical Infectious Diseases 2017 Pubmed
Plant Poly(A) Site and Ortholog Identification Algorithm and Data Visualization Research
- Li, Lei; Ji, Guoli; Ye, Congting; Shu, Changlong; Zhang, Jie; Liang, Chun; PlantOrDB: A genome-wide ortholog database for land plants and green algae BMC plant biology 2015 Pubmed
- Ji, Guoli; Li, Lei; Li, Qingshun Q; Wu, Xiangdong; Fu, Jingyi; Chen, Gong; Wu, Xiaohui; PASPA: a web server for mRNA poly (A) site predictions in plants and algae Bioinformatics 2015 Oxford Press
- Ye, Congting; Ji, Guoli; Li, Lei; Liang, Chun; detectIR: a novel program for detecting perfect and imperfect inverted repeats using complex numbers and vector calculation PloS one 2014 PLoS One
- Shi, Jieming; Dong, Min; Li, Lei; Liu, Lin; Luz-Madrigal, Agustin; Tsonis, Panagiotis A; Del Rio-Tsonis, Katia; Liang, Chun; mirPRo–a novel standalone program for differential expression and variation analysis of miRNAs Scientific reports 2015 Scientific Reports